吴萍副教授、蔡称心教授课题组在ACS SENSORS发表研究论文
Plasmonic Gold Nanohole Array for Surface-Enhanced Raman Scattering Detection of DNA Methylation
Luo, XJ (Luo, Xiaojun)[ 1 ] ; Xing, YF (Xing, Yingfang)[ 1 ] ; Galvan, DD (Galvan, Daniel David)[ 2 ] ; Zheng, EJ (Zheng, Erjin)[ 2 ] ; Wu, P (Wu, Ping)[ 1 ]*(吴萍); Cai, CX (Cai, Chenxin)[ 1 ]*(蔡称心); Yu, QM (Yu, Qiuming)[ 2 ]
[ 1 ] Nanjing Normal Univ, Coll Chem & Mat Sci, Jiangsu Collaborat Innovat Ctr Biomed Funct Mat, Jiangsu Key Lab New Power Batteries, Nanjing 210097, Jiangsu, Peoples R China
[ 2 ] Univ Washington, Dept Chem Engn, Seattle, WA 98195 USA
ACS SENSORS,201906,4(6),1534-1542
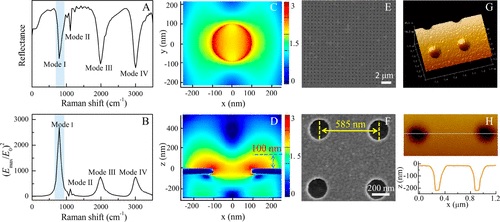
Surface-enhanced Raman spectroscopy (SERS), which utilizes nanogaps between noble-metal nanostructures as hot spots to yield ultrasensitive SERS signals, is an outstanding label-free and straightforward tool for DNA methylation analysis. Herein, a plasmonic gold nanohole array (PGNA) with well-controlled hot spots and an open surface was designed as a SERS substrate for DNA methylation detection. A finite-difference time-domain (FDTD) simulation was first employed to investigate the electric field distributions of the PGNA as a function of the geometric parameters. The plasmonic response was tuned to 785 cm-1 to match the ring breathing vibrational band of cytosine, the intensity change of which was revealed to be a marker of DNA methylation. Then, guided by the FDTD simulation results, the PGNA was fabricated via the electron beam lithography (EBL) technique. The fabricated PGNA had an open and easily accessible surface topology, a SERS enhancement factor of similar to 10(6), and a relative standard deviation (RSD) of 7.1% for 500 repetitions over an area of 20 x 20 mu m(2) using 1 mu M Rhodamine 6G as the Raman reporter. The fabricated PGNA was further used as a platform for determining DNA methylation. The proposed method exhibited a sensitivity for detecting 1% of methylation changes. Moreover, insight into the dynamic information on methylation events was obtained by combining principal component analysis (PCA) with 2D correlation spectroscopy analysis. Finally, clear discrimination of the different methylation sites, such as 5-methylcytosine and N6-methyladenine, was demonstrated.
文章链接:
https://pubs.acs.org/doi/10.1021/acssensors.9b00008
版权与免责声明:本网页的内容由收集互联网上公开发布的信息整理获得。目的在于传递信息及分享,并不意味着赞同其观点或证实其真实性,也不构成其他建议。仅提供交流平台,不为其版权负责。如涉及侵权,请联系我们及时修改或删除。邮箱:sales@allpeptide.com