400-998-5282
专注多肽 服务科研
400-998-5282
专注多肽 服务科研
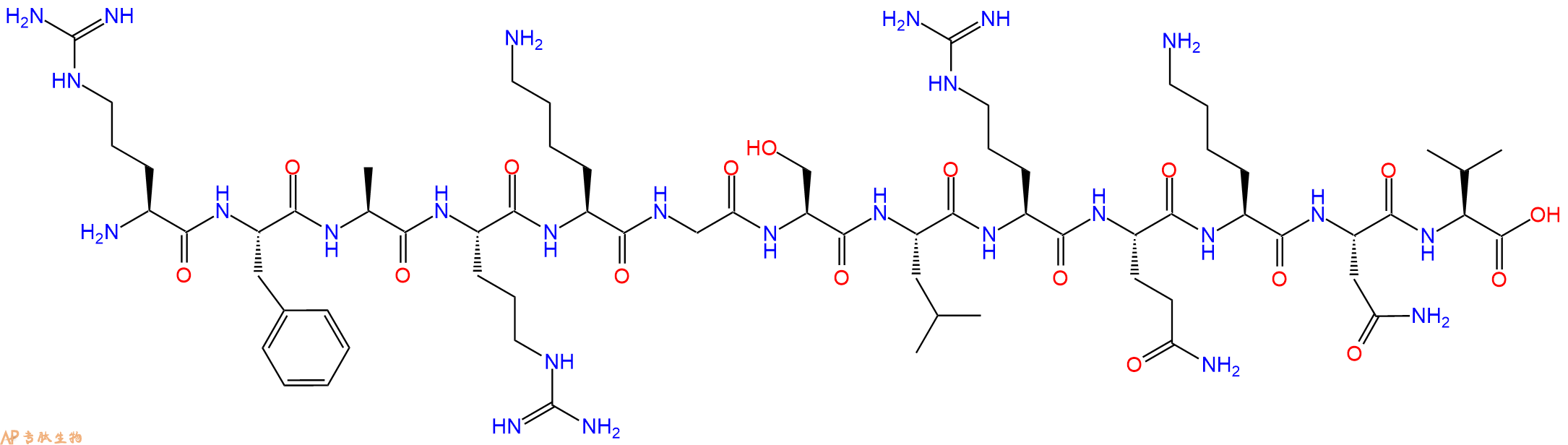
蛋白激酶C(19-31)是由PKCa(19-31)的伪底物调节结构域衍生而来的,第25位由丝氨酸取代了野生型PKCa的丙氨酸,常被用作蛋白激酶C的底物多肽来测定蛋白激酶C的活性。
编号:150352
CAS号:136795-05-6
单字母:H2N-RFARKGSLRQKNV-OH
| 参考文献(References): | C. House et al., Science, 238, 1726 (1987) |
高亲和力蛋白激酶C假底物。
High affinity protein kinase C pseudosubstrate.
背景
蛋白激酶C(19-31),化学式为C67H118N26O17,序列为H-ARG-PHE-ALA-ARG-LYS-GLY-SER-LEU-ARG-GLN-LYS-ASN-VAL-OH,分子量为1559.82。这种多肽是由PKCa(19-31)的伪底物调节结构域衍生而来的,第25位由丝氨酸取代了野生型PKCa的丙氨酸,常被用作蛋白激酶C的底物多肽来测定蛋白激酶C的活性。
蛋白激酶C也称为PKC,属于蛋白激酶家族,通过磷酸化其它蛋白质的丝氨酸和苏氨酸残基的羟基来调控蛋白的功能。研究表明,PKCa参与了许多不同的细胞过程,例如细胞粘附、细胞转化、细胞周期检测点和控制细胞体积。敲除小鼠的研究表明,该激酶可能是心脏收缩和肌细胞Ca2+处理的功能调节剂。
参考文献:
1. Mellor H, Parker PJ (1998). "The extended protein kinase C superfamily". Biochem. J.. 332 ( Pt 2): 281–92
2. Nishizuka Y (1995). "Protein kinase C and lipid signaling for sustained cellular responses" (abstract). FASEB J. 9 (7): 484–96.
3. Vicente Micol. Correlation between Protein Kinase C an Activity and Membrane Phase Behavior. Departamento de Bioquı´mica y Biologı´a Molecular
Caspase酶对应的底物,Caspases(半胱氨酸天冬氨酸蛋白酶,半胱氨酸依赖性天冬氨酸定向蛋白酶)是一类蛋白酶家族,其功能与凋亡(程序性细胞死亡),坏死和发烧(炎症)的过程密切相关。
什么是胱天蛋白酶?
胱天蛋白酶(Caspases)是含半胱氨酸的天冬氨酸蛋白水解酶,它们是为细胞凋亡的主要介质。多种受体,例如TNF-α 受体,FasL受体,TLR和死亡受体,以及Bcl-2和凋亡抑制剂(IAP)蛋白家族参与并调节该caspase依赖性凋亡途径。一旦Caspase受到上游信号(外部或内在)刺激被激活,即会参与执行下游蛋白底物的水解作用,并触发一系列事件,导致细胞分解,死亡,吞噬作用和细胞碎片的清除。
人Caspases酶
人的Caspases家族基于序列相似性和生物学功能等共性主要可分为三大类:第一类由具有长胱天蛋白酶募集结构域的“炎症”胱天蛋白酶组成,他们对P4位上的较大的芳香族或疏水性残基具有亲和力。第二类由具有短的前体结构域的“细胞凋亡效应”胱天蛋白酶组成,而第三类由具有长的前提结构域的Pap位置具有亮氨酸或缬氨酸底物亲和力的“凋亡引发剂”胱天蛋白酶组成(表1)。
表1. 人胱天蛋白酶的功能分类:
| 细胞死亡途径 | 半胱天冬酶类型 | 酵素 | 物种 |
| 细胞凋亡 | 启动器 | Caspases 2 | 人与鼠 |
| 细胞凋亡 | 启动器 | Caspases 8 | 人与鼠 |
| 细胞凋亡 | 启动器 | Caspases 9 | 人与鼠 |
| 细胞凋亡 | 启动器 | Caspases 10 | 人的 |
| 细胞凋亡 | 效应器 | Caspases 3 | 人与鼠 |
| 细胞凋亡 | 效应器 | Caspases 6 | 人与鼠 |
| 细胞凋亡 | 效应器 | Caspases 6 | 人与鼠 |
| 细胞焦亡 | 炎性的 | Caspases 1 | 人与鼠 |
| 细胞焦亡 | 炎性的 | Caspases 4 | 人的 |
| 细胞焦亡 | 炎性的 | Caspases 5 | 人的 |
启动器Caspase和效应器Caspase酶
根据其在凋亡胱天蛋白酶途径中的作用,胱天蛋白酶可分为两类:启动器和效应器Caspase酶。启动器和效应器Caspas酶都具有由小亚基和大亚基组成的催化位点,Caspase酶的识别位

凋亡启动器Caspase酶,例如caspase-2,-8,-9和-10可以启动caspase激活级联反应。Caspase-8对于形成死亡诱导信号复合物(DISC)是必不可少的,并且在激活后,Caspase-8激活下游效应子Caspase(例如Caspase 3)并介导线粒体中细胞色素c的释放。Caspase-8已被证明对IETD肽序列具有相对较高的底物选择性。凋亡效应胱天蛋白酶例如Caspase-3,-6和-7虽然不负责启动级联途径,但是当被激活时,它们在级联的中间和后续步骤中起着不可或缺的作用。Caspase-3(CPP32 / apopain)是关键效应器,因为它放大了来自启动器Caspase的信号,使用对Caspase-3有选择性的DEVD肽序列对活化的Caspase-3进行检测,可以检测Caspase-3的活性。
Caspase酶底物和抑制剂
Caspase底物和抑制剂由两个关键成分组成:Caspase识别序列和信号产生或蛋白酶抑制基序。不同Caspase识别序列不同,一般由三个或四个氨基酸组成(表2)。Caspase酶识别序列的N端通常有乙酰基(Ac)或碳苯甲氧基(Z)基团修饰,以增强膜的通透性。对应的Caspase识别特定的肽序列为其酶促反应切割位点,释放产生信号或抑制信号的基序。Caspase的显色和荧光底物均以相似的方式起作用,其中底物的信号或颜色强度与蛋白水解活性成正比。
表2. Caspase的底物及其序列
| 多肽 | 氨基酸序列 | 对应的Caspase的种类 |
| IETD | Ile-Glu-Thr-Asp | Caspase 8,颗粒酶B |
| DEVD | Asp-Glu-Val-Asp | Caspase 3、6、7、8或10 |
| LEHD | Leu-Glu-His-Asp | Caspase 9 |
| VAD | Val-Ala-Asp | Caspase 1、2、3、6、8、9或10 |
Caspase酶的显色底物
Caspase的显色底物是有Caspase识别序列及生色基团组成,常见的生色团有pNA(对硝基苯胺或4-硝基苯胺),可使用酶标仪或分光光度计在405 nm处进行光密度检测。
表3. Caspase的显色底物
| 底物 | Caspase | 吸收(nm) | 颜色 |
| Ac-DEVD-pNA * CAS 189950-66-1 * | 半胱天冬酶3 | 405 nm | 黄色 |
| Z-DEVD-pNA | 半胱天冬酶3 | 405 nm | 黄色 |
| Z-IETD-pNA * CAS 219138-21-3 * | 半胱天冬酶8,颗粒酶B | 405 nm | 黄色 |
Caspase的荧光底物
Caspase的荧光底物的结构包含与半胱天冬酶识别相关的荧光团,例如7-氨基-4-甲基香豆素(AMC),7-氨基-4-三氟甲基香豆素(AFC), Rhodamine 110(R110)或ProRed™620。R110的Caspase底物比基于香豆素的Caspase底物(例如AMC和AFC)更敏感,但由于两步裂解过程,其动态范围更窄。 建议将R110标记的Caspase底物用于终点法测定,而将AMC和AFC标记的 Caspase底物用于动力学测定。

图.从左到右,分别是AMC(7-氨基-4-甲基香豆素),AFC(7-氨基-4-三氟甲基香豆素),Rhodamine 110(R110)和ProRed™620的激发和发射光谱。
表4.荧光半胱天冬酶底物。
| 底物名称 | 对应的Caspase | Ex(nm) | Em(nm) | ε¹ | Φ² |
| Ac-DEVD-AFC * CAS 201608-14-2 * | 半胱天冬酶3、7 | 376 | 482 | 17000 | 0.53 |
| Ac-DEVD-AMC * CAS 169332-61-0 * | 半胱天冬酶3、7 | 341 | 441 | 19000 | N / D |
| Z-DEVD-AFC | 半胱天冬酶3、7 | 376 | 482 | 17000 | 0.53 |
| Z-DEVD-AMC * CAS 1135416-11-3 * | 半胱天冬酶3、7 | 341 | 441 | 19000 | N / D |
| Z-DEVD-ProRed™620 | 半胱天冬酶3、7 | 532 | 619 | N / D | N / D |
| (Z-DEVD)2 -R110 * CAS 223538-61-2 * | 半胱天冬酶3、7 | 500 | 522 | 80000 | N / D |
| Z-DEVD-ProRed™620 | 半胱天冬酶3、7 | 532 | 619 | N / D | N / D |
| Ac-IETD-AFC * CAS 211990-57-7 * | 半胱天冬酶8,颗粒酶B | 376 | 482 | 17000 | 0.53 |
| Z-IETD-AFC * CAS 219138-02-0 * | 半胱天冬酶8,颗粒酶B | 376 | 482 | 17000 | 0.53 |
注意:
1.ε=在其最大吸收波长处的摩尔消光系数(单位= cm -1M -1)。
2.Φ=水性缓冲液(pH 7.2)中的荧光量子产率。
Caspase抑制剂
Caspase抑制剂能与Caspase的活性位点结合并形成可逆或不可逆的连接,通常,Caspase抑制剂的结构由Caspase识别序列,诸如醛(-CHO)或氟甲基酮(-FMK)的官能团组成。具有醛官能团的胱天蛋白酶抑制剂是可逆的,而具有FMK的抑制剂是不可逆的。半胱天冬酶底物和抑制剂都具有较小的细胞毒性作用,因此,它们是研究半胱天冬酶活性的有用工具。
表5. 可逆和不可逆的Caspase酶抑制剂
| 抑制剂 | Caspase的种类 | 是否可逆 | Ex(nm) | Em(nm) |
| Ac-DEVD-CHO * CAS 169332-60-9 * | 半胱天冬酶3、7 | 可逆的 | -- | -- |
| Ac-IETD-CHO * CAS 191338-86-0 * | 半胱天冬酶8 | 可逆的 | -- | -- |
| mFluor™450-VAD-FMK | 半胱天冬酶1,2,3,6,8,9,10 | 不可逆的 | 406 | 445 |
| mFluor™510-VAD-FMK | 半胱天冬酶1,2,3,6,8,9,10 | 不可逆的 | 412 | 505 |
| FITC-C6-DEVD-FMK | 半胱天冬酶3、7 | 不可逆的 | 491 | 516 |
| FITC-C6-DEVD-FMK | 半胱天冬酶3、7 | 不可逆的 | 491 | 516 |
| FITC-C6-LEHD-FMK | 半胱天冬酶9 | 不可逆的 | 491 | 516 |
| FITC-C6-LEHD-FMK | 半胱天冬酶9 | 不可逆的 | 491 | 516 |
| FAM-VAD-FMK | 半胱天冬酶1,2,3,6,8,9,10 | 不可逆的 | 493 | 517 |
| SRB-VAD-FMK [磺胺丁胺B-VAD-FMK] | 半胱天冬酶1,2,3,6,8,9,10 | 不可逆的 | 559 | 577 |
Definition
Protein kinases are transferase that catalyze the phosphorylation of proteins by covalently attaching phosphate groups to them, using ATP as a phosphate donor. Reversible protein phosphorylation-dephosphorylation has a principal role in the regulation of essentially all cellular functions. Kinase/phosphatase substrates can be found grouped according to their kinase families. A single substrate can have a many number of modifications.
Discovery
Phoebus AL at the Rockefeller Institute identified phosphate in the protein Vitellin (phosvitin) in 1906 1 and by 1933 Fritz Lipmann had detected phosphoserine in Casein 2. In 1954 Eugene P. Kennedy described the first ‘enzymatic phosphorylation of proteins’ in a variety of normal and malignant tissues, he showed that the phosphorus of the phosphoprotein fraction undergoes a high rate of turnover, as measured by incorporation of 32P 3 .
Structural Characteristics
There are thousands of different kinds of proteins in any particular cell that are substrate for different kinases and phosphatases. Phosphorylation of any site on a given protein can change the structure, function or localization of that protein. Within a protein, phosphorylation can occur on several amino acids. Phosphorylation on serine is the most common, followed by threonine. Tyrosine phosphorylation is relatively rare. However, since tyrosine phosphorylated proteins are relatively easy to purify using antibodies, tyrosine phosphorylation sites are relatively well understood. Histidine and aspartate phosphorylation occurs in prokaryotes as part of two-component signaling and in some cases in eukaryotes in some signal transduction pathways 4. Phosphorylation of seryl or threonyl (and occasionally tyrosyl) residues triggers small conformational changes in these proteins that alter their biological properties.
Mode of Action
Phosphatase removes a phosphate group from its substrate by hydrolysing phosphoric acid monoesters into a phosphate ion and a molecule with a free hydroxyl group. Protein kinases are the effectors of phosphorylation and catalyse the transfer of a y-phosphate from ATP to specific amino acids on proteins. The addition of a phosphate (PO4) molecule to a polar R group of an amino acid residue can turn a hydrophobic portion of a protein into a polar and extremely hydrophilic portion of molecule. In this way it can introduce a conformational change in the structure of the protein via interaction with other hydrophobic and hydrophilic residues in the protein. Several protein kinases are important in cellular control (e.g. glycogen synthase kinase-3, acetyl CoA carboxylase kinase, tyrosine hydroxylase kinase and casein kinase-2), which are themselves controlled by allosteric effectors, phosphorylation, insulin and other growth factors, or by regulators. Protein phosphatase catalytic units are responsible for dephosphorylating many regulated proteins in the cytoplasm that are phosphorylated on serine and threonine residues. Some protein phosphatases are controlled by second messengers. PP-1(Protein phosphatase-1) is regulated by cyclic AMP in several ways that vary with the form of the enzyme and the tissue. It is inhibited by cyclic AMP through the phosphorylation of inhibitor-1 and its isoforms through the phosphorylation of targeting proteins such as the glycogen-binding subunit, and through allosteric inhibition by phosphorylase a. PP-2B (Protein phosphatase-2B) is activated by Ca2+ through the interaction of this second messenger with an integral Ca2+ -binding subunit, as well as calmodulin itself. Protein phosphorylation-dephosphorylation is the basis of a network of interlocking systems that allow hormones and other extracellular signals, acting through just a few second messengers, to coordinate biochemical functions 5.
Functions
Reversible phosphorylation of proteins is an important regulatory mechanism that occurs in living cells 6. Reversible phosphorylation results in a change in conformation the structure in many enzymes and receptors, causing them to become activated or deactivated.
Regulatory roles, the p53 protein is heavily regulated through phosphorylation sites, it has 18 different phosphorylation sites. Activation and phosphorylation of p53 can lead to cell cycle arrest, which can be reversed under some circumstances, or apoptotic cell death 7. In energy-requiring reactions, phosphorylation of Na+/K+-ATPase during the transport of sodium (Na+) and potassium(K+) ions across the cell membrane in osmoregulation to maintain homeostasis.
Enzyme regulation, phosphorylation of the enzyme GSK-3 by AKT (Protein kinase B) is a important regulation in insulin signaling pathway.
Protein-protein interaction, phosphorylation of the cytosolic components of NADPH oxidase, a large membrane-bound, multi-protein enzyme plays an important role in the regulation of protein-protein interactions of the enzyme.
Protein degradation, phosphorylation of some proteins causes them to be degraded by the ATP-dependent ubiquitin/proteasome pathway. These proteins become substrates for particular E3 ubiquitin ligases only when they are phosphorylated 8.
References
1. Levene PA, Alsberg CL (1906). The cleavage products of vitellin. J. Biol. Chem., 2(1): 127-133.
2. Lipmann FA, Levene PA (1932). Serinephosphoric acid obtained on hydrolysis of vitellinic acid. J. Biol. Chem., 98 (1):109-114.
3. Burnett G, Kennedy EP (1954). The enzymatic phosphorylation of proteins. J. Biol. Chem., 211(2):969–980.
4. Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, Edgar D, Ekblom P, Engel J, Engvall E, Hohenester E, Jones JC, Kleinman HK, Marinkovich MP, Martin GR, Mayer U, Meneguzzi G, Miner JH, Miyazaki K, Patarroyo M, Paulsson M, Quaranta V, Sanes JR, Sasaki T, Sekiguchi K, Sorokin LM, Talts JF, Tryggvason K, Uitto J, Virtanen I, von der Mark K, Wewer UM, Yamada Y, Yurchenco PD (2005). A simplified laminin nomenclature. Matrix Biol., 24(5):326-332.
5. Cohen P (1988). Protein Phosphorylation and Hormone Action. Proceedings of the Royal Society of London. Biological Sciences, 234(1275):115-144.
6. Barford D, Das AK, Egloff MP (1998). The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct., 27:133–164.
7. Ashcroft M, Kubbutat MH, Vousden KH (1999). Regulation of p53 function and stability by phosphorylation. Mol. Cell. Biol., 19(3):1751–1758.
8. Babior BM (1999). NADPH oxidase: an update. Blood, 93(5):1464–1476.





