400-998-5282
专注多肽 服务科研
400-998-5282
专注多肽 服务科研
可显着逆转小鼠低水平的吗啡镇痛感受性。
编号:156270
CAS号:121932-06-7
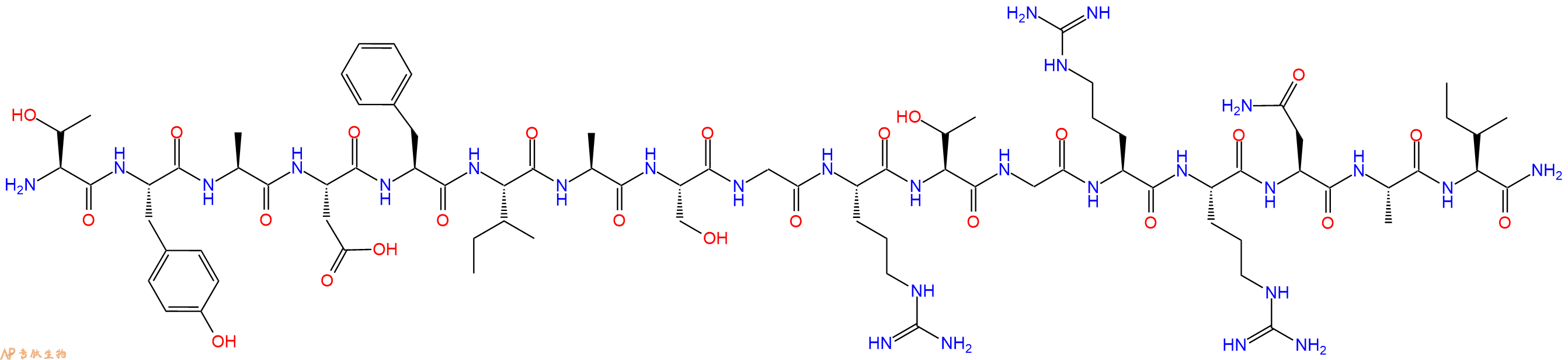
单字母:H2N-TYADFIASGRTGRRNAI-CONH2
PKA Inhibitor Fragment (6-22) amide 是 cAMP 依赖性蛋白激酶 A (PKA) 的抑制剂,Ki 值为 2.8 nM。PKA Inhibitor Fragment (6-22) amide 可显着逆转小鼠低水平的吗啡镇痛感受性。
PKA Inhibitor Fragment (6-22) amide is an inhibitor of cAMP-dependent protein kinase A (PKA), with a Ki of 2.8 nM. PKA Inhibitor Fragment (6-22) amide can significantly reverse low-level morphine antinociceptive tolerance in mice.
PKA抑制剂片段(6-22)酰胺乙酸酯是cAMP依赖性蛋白激酶(PKA)(Ki=2.5nM)的合成肽抑制剂,来源于热稳定的PKA抑制剂蛋白PKI。它是最短的合成PKI肽,对PKA的抑制具有很高的效力。
PKA Inhibitor Fragment (6-22) amide acetate is a synthetic peptide inhibitor of cAMP-dependent protein kinase (PKA) (Ki = 2.5 nM), derived from the heat-stable PKA inhibitor protein PKI. It is the shortest synthetic PKI peptide that retains high potency for PKA inhibition.
Katz BM, et, al. Synthesis, characterization and inhibitory activities of (4-N3[3,5-3H]Phe10)PKI(6-22)amide and its precursors: photoaffinity labeling peptides for the active site of cyclic AMP-dependent protein kinase. Int J Pept Protein Res. 1989 Jun;33(6):439 : https://pubmed.ncbi.nlm.nih.gov/2550380/
Dalton GD, et, al. Alterations in brain Protein Kinase A activity and reversal of morphine tolerance by two fragments of native Protein Kinase A inhibitor peptide (PKI). Neuropharmacology. 2005 Apr; 48(5): 648-57. : https://pubmed.ncbi.nlm.nih.gov/15814100/
多肽H2N-Thr-Tyr-Ala-Asp-Phe-Ile-Ala-Ser-Gly-Arg-Thr-Gly-Arg-Arg-Asn-Ala-Ile-NH2的合成步骤:
1、合成MBHA树脂:取若干克的MBHA树脂(如初始取代度为0.5mmol/g)和1倍树脂摩尔量的Fmoc-Linker-OH加入到反应器中,加入DMF,搅拌使氨基酸完全溶解。再加入树脂2倍量的DIEPA,搅拌混合均匀。再加入树脂0.95倍量的HBTU,搅拌混合均匀。反应3-4小时后,用DMF洗涤3次。用2倍树脂体积的10%乙酸酐/DMF 进行封端30分钟。然后再用DMF洗涤3次,甲醇洗涤2次,DCM洗涤2次,再用甲醇洗涤2次。真空干燥12小时以上,得到干燥的树脂{Fmoc-Linker-MHBA Resin},测定取代度。这里测得取代度为 0.3mmol/g。结构如下图:
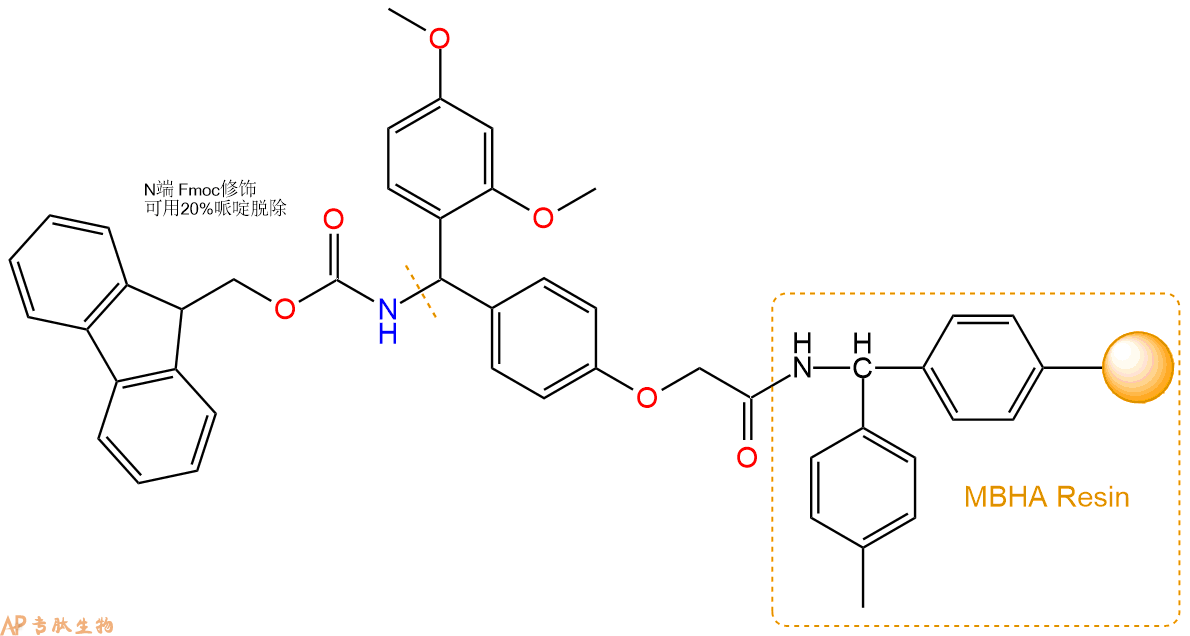
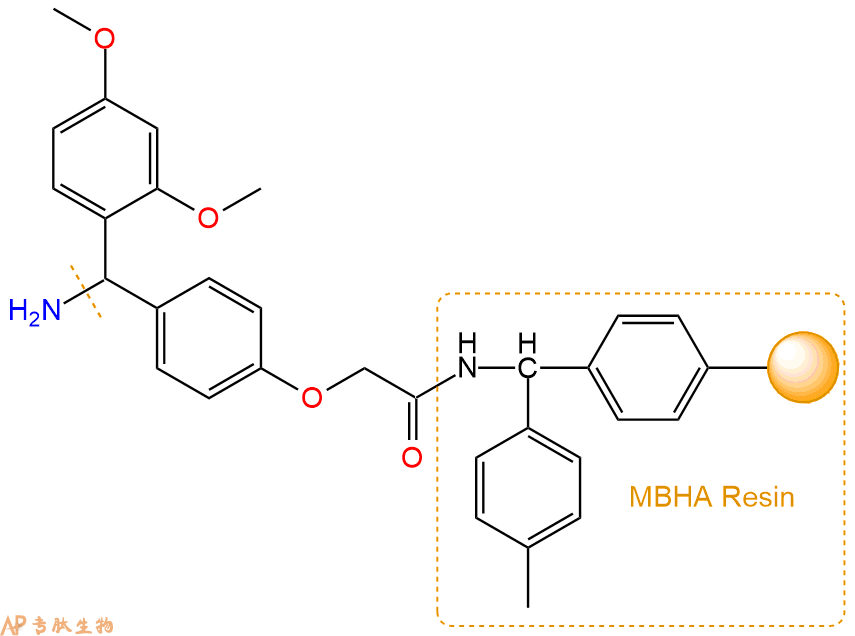
2、脱Fmoc:取2.67g的上述树脂,用DCM或DMF溶胀20分钟。用DMF洗涤2遍。加3倍树脂体积的20%Pip/DMF溶液,鼓氮气30分钟,然后2倍树脂体积的DMF 洗涤5次。得到 H2N-Linker-MBHA Resin 。(此步骤脱除Fmoc基团,茚三酮检测为蓝色,Pip为哌啶)。结构图如下:
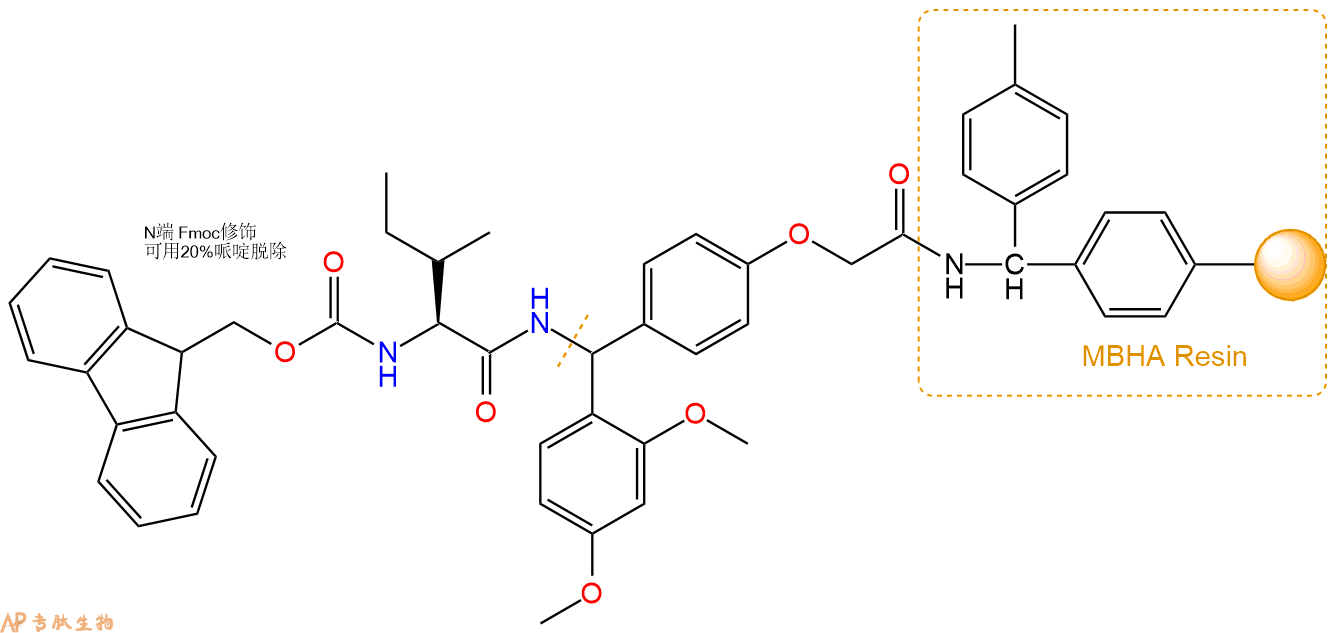
3、缩合:取2.4mmol Fmoc-Ile-OH 氨基酸,加入到上述树脂里,加适当DMF溶解氨基酸,再依次加入4.81mmol DIPEA,2.28mmol HBTU。反应30分钟后,取小样洗涤,茚三酮检测为无色。用2倍树脂体积的DMF 洗涤3次树脂。(洗涤树脂,去掉残留溶剂,为下一步反应做准备)。得到Fmoc-Ile-Linker-MBHA Resin。氨基酸:DIPEA:HBTU:树脂=3:6:2.85:1(摩尔比)。结构图如下:
4、依次循环步骤二、步骤三,依次得到
H2N-Ile-Linker-MBHA Resin
Fmoc-Ala-Ile-Linker-MBHA Resin
H2N-Ala-Ile-Linker-MBHA Resin
Fmoc-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Phe-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Phe-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Asp(OtBu)-Phe-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Asp(OtBu)-Phe-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Ala-Asp(OtBu)-Phe-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Ala-Asp(OtBu)-Phe-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Tyr(tBu)-Ala-Asp(OtBu)-Phe-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
H2N-Tyr(tBu)-Ala-Asp(OtBu)-Phe-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
Fmoc-Thr(tBu)-Tyr(tBu)-Ala-Asp(OtBu)-Phe-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin
以上中间结构,均可在专肽生物多肽计算器-多肽结构计算器中,一键画出。
最后再经过步骤二得到 H2N-Thr(tBu)-Tyr(tBu)-Ala-Asp(OtBu)-Phe-Ile-Ala-Ser(tBu)-Gly-Arg(Pbf)-Thr(tBu)-Gly-Arg(Pbf)-Arg(Pbf)-Asn(Trt)-Ala-Ile-Linker-MBHA Resin,结构如下:
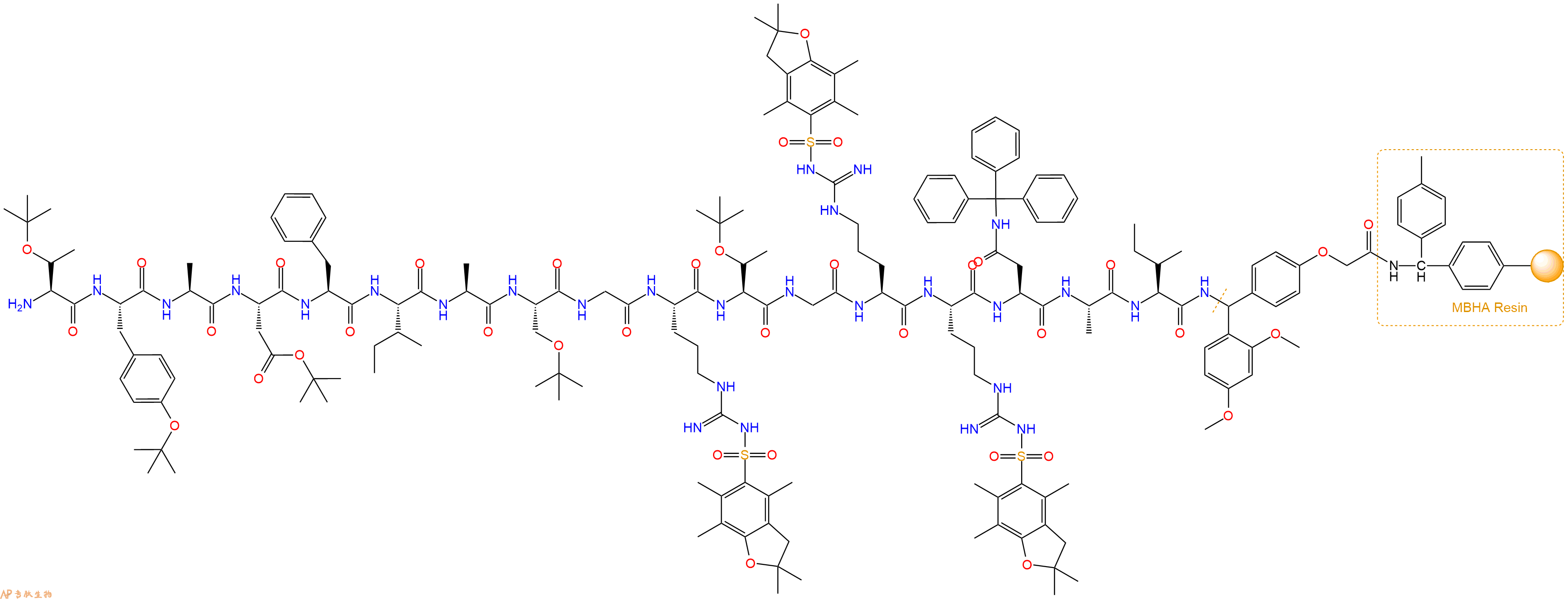
5、切割:6倍树脂体积的切割液(或每1g树脂加8ml左右的切割液),摇床摇晃 2小时,过滤掉树脂,用冰无水乙醚沉淀滤液,并用冰无水乙醚洗涤沉淀物3次,最后将沉淀物放真空干燥釜中,常温干燥24小试,得到粗品H2N-Thr-Tyr-Ala-Asp-Phe-Ile-Ala-Ser-Gly-Arg-Thr-Gly-Arg-Arg-Asn-Ala-Ile-NH2。结构图见产品结构图。
切割液选择:1)TFA:H2O=95%:5%
2)TFA:H2O:TIS=95%:2.5%:2.5%
3)三氟乙酸:茴香硫醚:1,2-乙二硫醇:苯酚:水=87.5%:5%:2.5%:2.5%:2.5%
(前两种适合没有容易氧化的氨基酸,例如Trp、Cys、Met。第三种适合几乎所有的序列。)
6、纯化冻干:使用液相色谱纯化,收集目标峰液体,进行冻干,获得蓬松的粉末状固体多肽。不过这时要取小样复测下纯度 是否目标纯度。
7、最后总结:
杭州专肽生物技术有限公司(ALLPEPTIDE https://www.allpeptide.com)主营定制多肽合成业务,提供各类长肽,短肽,环肽,提供各类修饰肽,如:荧光标记修饰(CY3、CY5、CY5.5、CY7、FAM、FITC、Rhodamine B、TAMRA等),功能基团修饰肽(叠氮、炔基、DBCO、DOTA、NOTA等),同位素标记肽(N15、C13),订书肽(Stapled Peptide),脂肪酸修饰肽(Pal、Myr、Ste),磷酸化修饰肽(P-Ser、P-Thr、P-Tyr),环肽(酰胺键环肽、一对或者多对二硫键环),生物素标记肽,PEG修饰肽,甲基化修饰肽等。
以上所有内容,为专肽生物原创内容,请勿发布到其他网站上。





