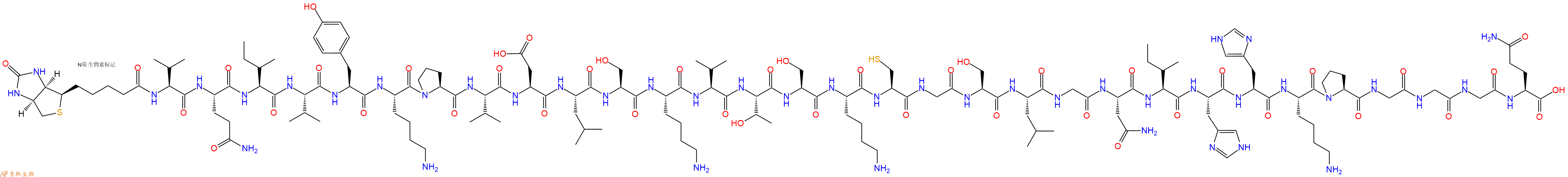
编号:130117
CAS号:2022956-58-5
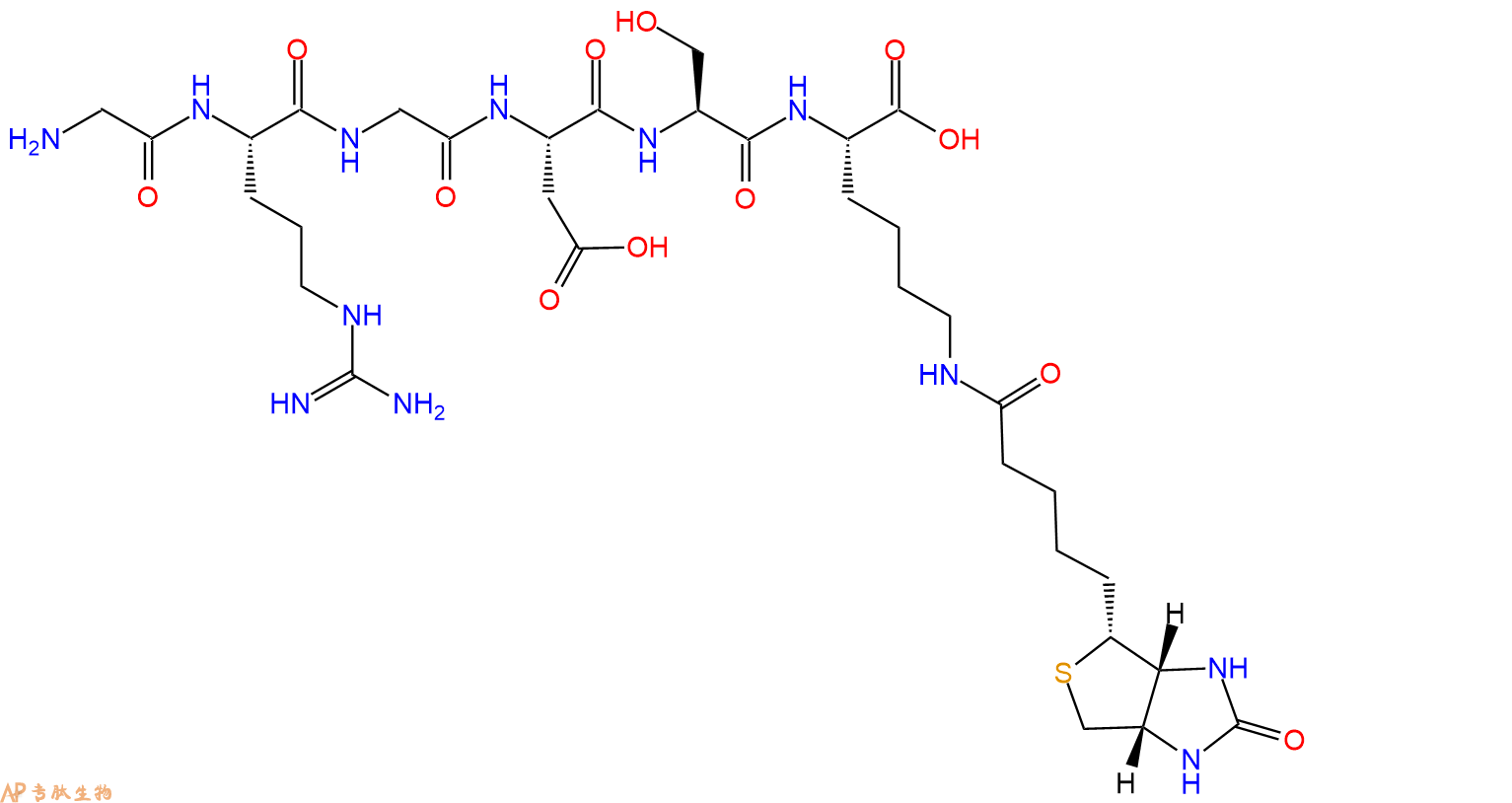
单字母:Biotinyl-VQIVYKPVDLSKVTSKCGSLGNIHHKPGGGQ-OH
| 编号: | 130117 |
| 中文名称: | 生物素标记肽Biotin-VQIVYKPVDLSKVTSKCGSLGNIHHKPGGGQ |
| 英文名: | Biotin-Val-Gln-Ile-Val-Tyr-Lys-Pro-Val-Asp-Leu-Ser |
| CAS号: | 2022956-58-5 |
| 单字母: | Biotinyl-VQIVYKPVDLSKVTSKCGSLGNIHHKPGGGQ-OH |
| 三字母: | Biotinyl N端生物素标记 -Val缬氨酸 -Gln谷氨酰胺 -Ile异亮氨酸 -Val缬氨酸 -Tyr酪氨酸 -Lys赖氨酸 -Pro脯氨酸 -Val缬氨酸 -Asp天冬氨酸 -Leu亮氨酸 -Ser丝氨酸 -Lys赖氨酸 -Val缬氨酸 -Thr苏氨酸 -Ser丝氨酸 -Lys赖氨酸 -Cys半胱氨酸 -Gly甘氨酸 -Ser丝氨酸 -Leu亮氨酸 -Gly甘氨酸 -Asn天冬酰胺 -Ile异亮氨酸 -His组氨酸 -His组氨酸 -Lys赖氨酸 -Pro脯氨酸 -Gly甘氨酸 -Gly甘氨酸 -Gly甘氨酸 -Gln谷氨酰胺 -OHC端羧基 |
| 氨基酸个数: | 31 |
| 分子式: | C153H250N44O44S2 |
| 平均分子量: | 3474.02 |
| 精确分子量: | 3471.81 |
| 等电点(PI): | 11.32 |
| pH=7.0时的净电荷数: | 5.44 |
| 平均亲水性: | -0.10416666666667 |
| 疏水性值: | -0.3 |
| 消光系数: | 1490 |
| 来源: | 人工化学合成,仅限科学研究使用,不得用于人体。 |
| 盐体系: | 可选TFA、HAc、HCl或其它 |
| 储存条件: | 负80℃至负20℃ |
| 标签: | 生物素标记肽(Biotinyl) Tau肽 |
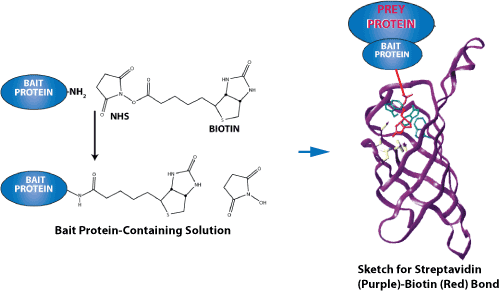
专肽生物合成用于蛋白质-蛋白质相互作用研究的生物素化肽。尽管生物素可以在 N 端或 C 端引入(通过赖氨酸残基),但我们建议使用 N 端修饰,因为它成本低、成功率高、周转时间短且易于操作。因为多肽合成是从 C 端到 N 端合成的,因此,N 端修饰是 SPPS步骤的最后一步,不需要额外的特定缩合步骤。相比之下,C 端修饰需要额外的步骤,并且通常更复杂。当然,原则上生物素可以定位在任何地方。

生物素可以通过多种不同的接头或间隔物与肽分离。尽管如此,还是建议包含一个灵活的间隔物,例如 Ahx(一个 6 碳接头),以使生物素标签更加稳定或灵活。
专肽生物在 N 端或 C 端提供生物素化:生物素-N 端、赖氨酸-生物素-肽中间和赖氨酸-生物素-C 端。
专肽生物还可以使用 Ahx 接头或长碳 (LC) 接头提供生物素化:生物素-Ahx-N 末端、Lys-Ahx-生物素-肽中间、Lys-Ahx-生物素-C-末端。
(生物素结构)
示例:
GRGDS在N端和C端标记生物素的结构展示。
1、GRGDS在N端标记生物素,不增加Ahx 接头
2、GRGDS在N端标记生物素,增加一个Ahx 接头

3、GRGDS在C端标记生物素,不增加Ahx 接头
4、GRGDS在C端标记生物素,增加一个Ahx 接头。
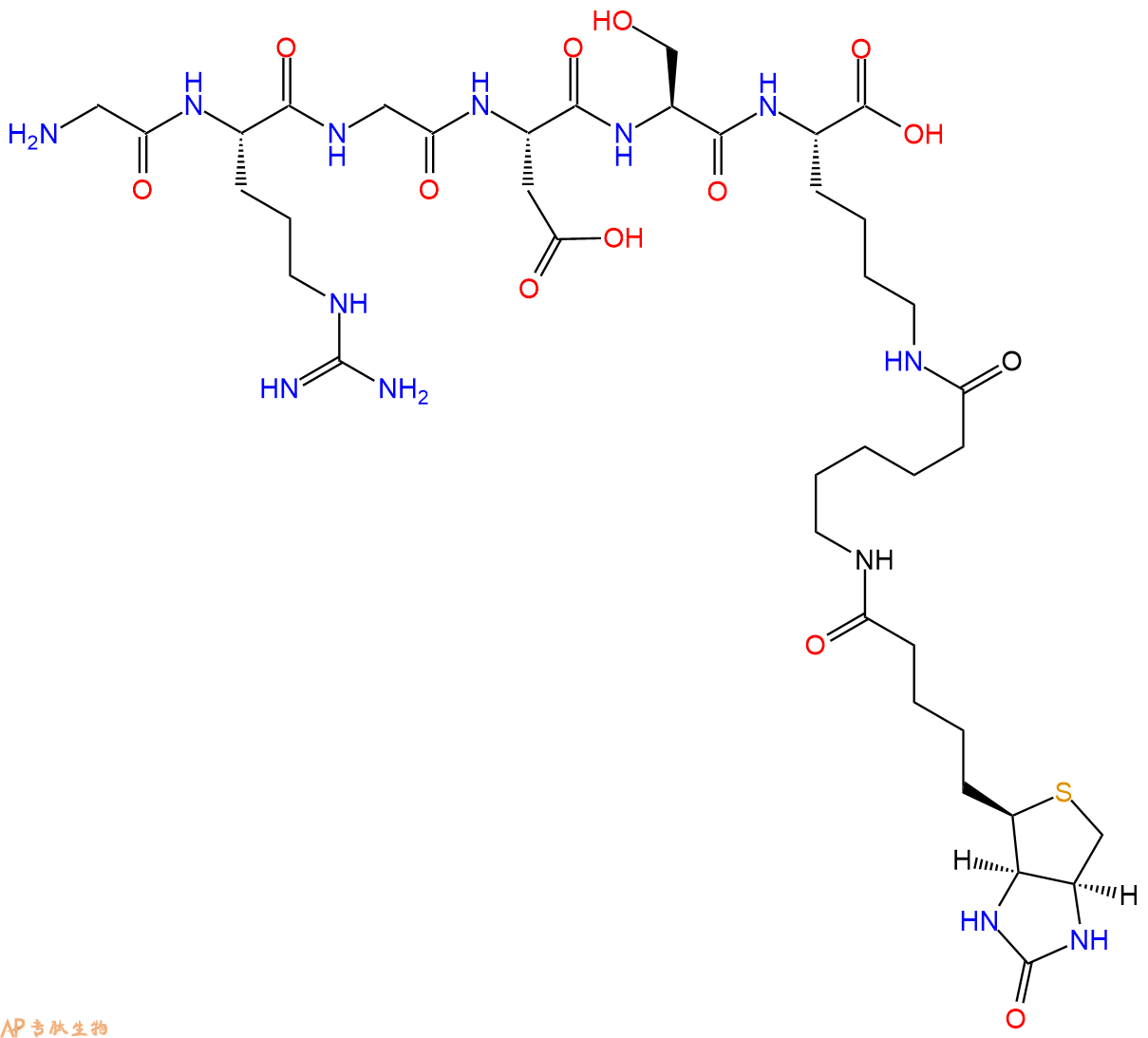
The amphipathic helical structure of the third fragment (306-336), in the four-repeat microtubule-binding domain of tau, is hypothesized to be responsible for the formation of the neuropathological filament. The additional biotin moiety will bind avidin or streptavidin with high specificity and affinity, providing robust results.
多肽Biotin-Val-Gln-Ile-Val-Tyr-Lys-Pro-Val-Asp-Leu-Ser-Lys-Val-Thr-Ser-Lys-Cys-Gly-Ser-Leu-Gly-Asn-Ile-His-His-Lys-Pro-Gly-Gly-Gly-Gln-COOH的合成步骤:
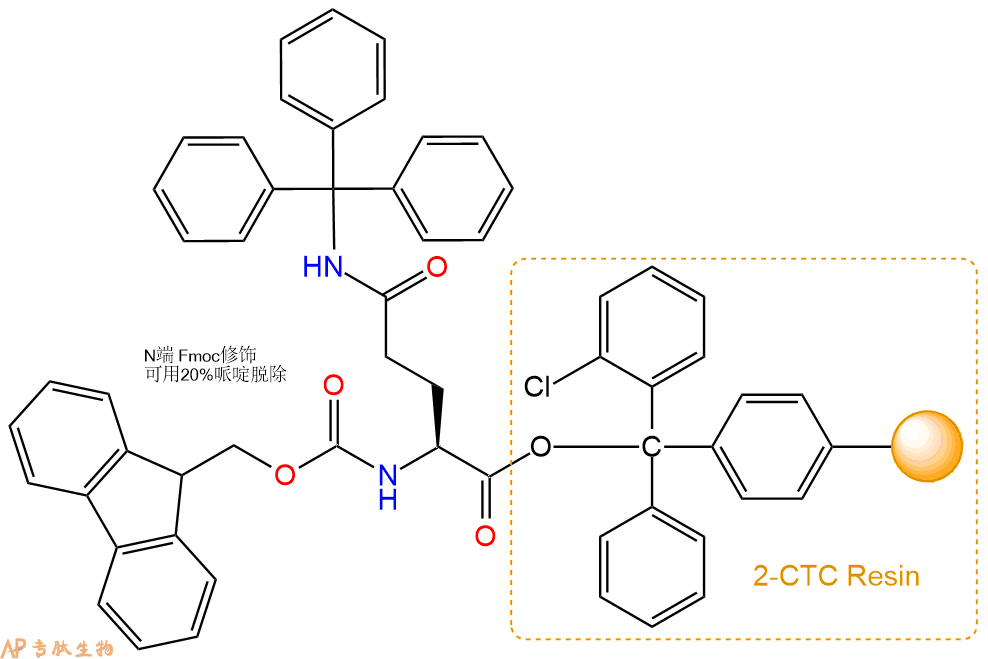
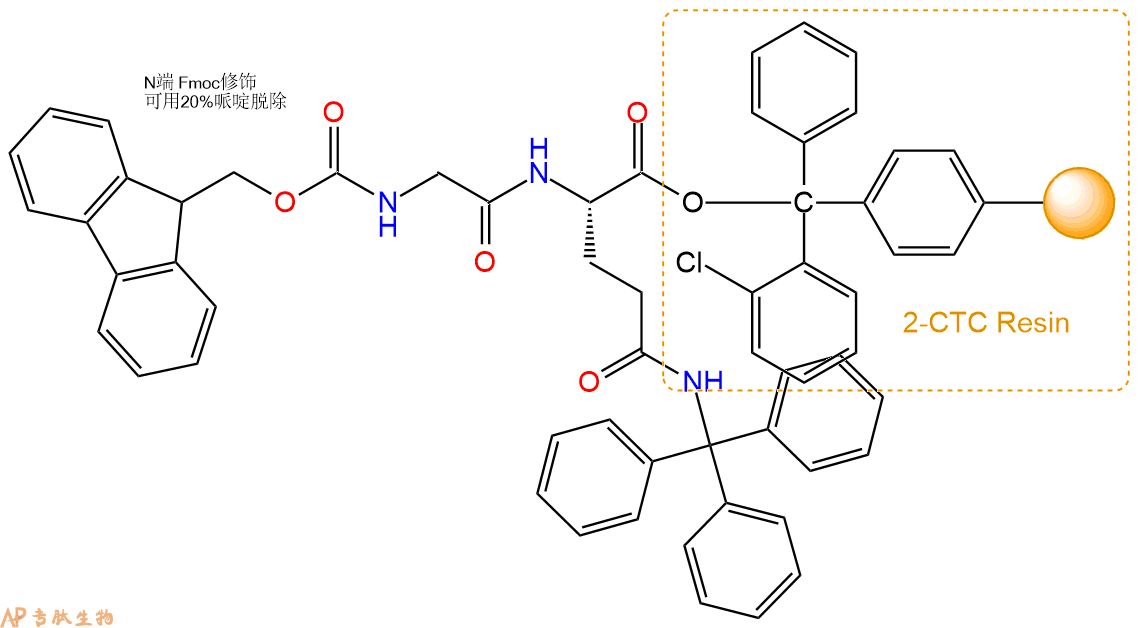
1、合成CTC树脂:称取0.18g CTC Resin(如初始取代度约为0.83mmol/g)和0.18mmol Fmoc-Gln(Trt)-OH于反应器中,加入适量DCM溶解氨基酸(需要注意,此时CTC树脂体积会增大好几倍,避免DCM溶液过少),再加入0.45mmol DIPEA(Mw:129.1,d:0.740g/ml),反应2-3小时后,可不抽滤溶液,直接加入1ml的HPLC级甲醇,封端半小时。依次用DMF洗涤2次,甲醇洗涤1次,DCM洗涤一次,甲醇洗涤一次,DCM洗涤一次,DMF洗涤2次(这里使用甲醇和DCM交替洗涤,是为了更好地去除其他溶质,有利于后续反应)。得到 Fmoc-Gln(Trt)-CTC Resin。结构图如下:
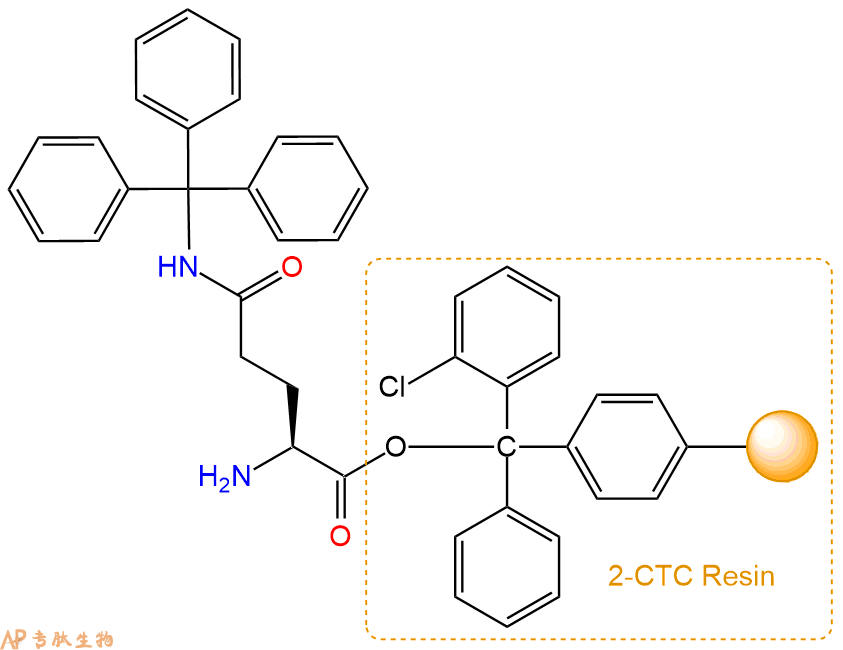
2、脱Fmoc:加3倍树脂体积的20%Pip/DMF溶液,鼓氮气30分钟,然后2倍树脂体积的DMF 洗涤5次。得到 H2N-Gln(Trt)-CTC Resin 。(此步骤脱除Fmoc基团,茚三酮检测为蓝色,Pip为哌啶)。结构图如下:
3、缩合:取0.45mmol Fmoc-Gly-OH 氨基酸,加入到上述树脂里,加适当DMF溶解氨基酸,再依次加入0.9mmol DIPEA,0.43mmol HBTU。反应30分钟后,取小样洗涤,茚三酮检测为无色。用2倍树脂体积的DMF 洗涤3次树脂。(洗涤树脂,去掉残留溶剂,为下一步反应做准备)。得到Fmoc-Gly-Gln(Trt)-CTC Resin。氨基酸:DIPEA:HBTU:树脂=3:6:2.85:1(摩尔比)。结构图如下:
4、依次循环步骤二、步骤三,依次得到
H2N-Gly-Gln(Trt)-CTC Resin
Fmoc-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Tyr(tBu)-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Tyr(tBu)-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Val-Tyr(tBu)-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Val-Tyr(tBu)-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Ile-Val-Tyr(tBu)-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Ile-Val-Tyr(tBu)-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Gln(Trt)-Ile-Val-Tyr(tBu)-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
H2N-Gln(Trt)-Ile-Val-Tyr(tBu)-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
Fmoc-Val-Gln(Trt)-Ile-Val-Tyr(tBu)-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin
以上中间结构,均可在专肽生物多肽计算器-多肽结构计算器中,一键画出。
最后再经过步骤二得到 H2N-Val-Gln(Trt)-Ile-Val-Tyr(tBu)-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTC Resin,结构如下:

5、生物素反应连接:在上述树脂中,加入适当DMF后,再加入0.45mmol 生物素到树脂中,再加入0.9mmol DIPEA、0.43mmol HBTU,鼓氮气反应30分钟。用2倍树脂体积的DMF 洗涤3次树脂(洗涤树脂,去掉残留溶剂,为下一步反应做准备)。 得到Biotin-Val-Gln(Trt)-Ile-Val-Tyr(tBu)-Lys(Boc)-Pro-Val-Asp(OtBu)-Leu-Ser(tBu)-Lys(Boc)-Val-Thr(tBu)-Ser(tBu)-Lys(Boc)-Cys(Trt)-Gly-Ser(tBu)-Leu-Gly-Asn(Trt)-Ile-His(Trt)-His(Trt)-Lys(Boc)-Pro-Gly-Gly-Gly-Gln(Trt)-CTCResin。 结构如下:
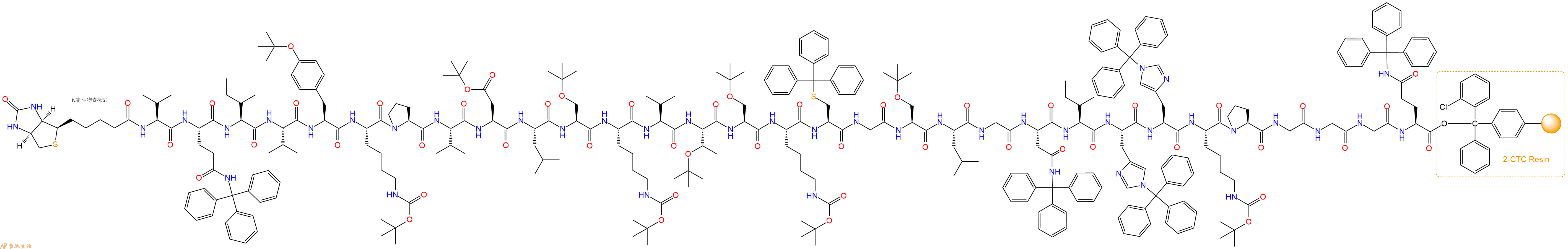
6、切割:6倍树脂体积的切割液(或每1g树脂加8ml左右的切割液),摇床摇晃 2小时,过滤掉树脂,用冰无水乙醚沉淀滤液,并用冰无水乙醚洗涤沉淀物3次,最后将沉淀物放真空干燥釜中,常温干燥24小试,得到粗品Biotin-Val-Gln-Ile-Val-Tyr-Lys-Pro-Val-Asp-Leu-Ser-Lys-Val-Thr-Ser-Lys-Cys-Gly-Ser-Leu-Gly-Asn-Ile-His-His-Lys-Pro-Gly-Gly-Gly-Gln-COOH。结构图见产品结构图。
切割液选择:1)TFA:H2O=95%:5%
2)TFA:H2O:TIS=95%:2.5%:2.5%
3)三氟乙酸:茴香硫醚:1,2-乙二硫醇:苯酚:水=87.5%:5%:2.5%:2.5%:2.5%
(前两种适合没有容易氧化的氨基酸,例如Trp、Cys、Met。第三种适合几乎所有的序列。)
6、纯化冻干:使用液相色谱纯化,收集目标峰液体,进行冻干,获得蓬松的粉末状固体多肽。不过这时要取小样复测下纯度 是否目标纯度。
7、最后总结:
杭州专肽生物技术有限公司(ALLPEPTIDE https://www.allpeptide.com)主营定制多肽合成业务,提供各类长肽,短肽,环肽,提供各类修饰肽,如:荧光标记修饰(CY3、CY5、CY5.5、CY7、FAM、FITC、Rhodamine B、TAMRA等),功能基团修饰肽(叠氮、炔基、DBCO、DOTA、NOTA等),同位素标记肽(N15、C13),订书肽(Stapled Peptide),脂肪酸修饰肽(Pal、Myr、Ste),磷酸化修饰肽(P-Ser、P-Thr、P-Tyr),环肽(酰胺键环肽、一对或者多对二硫键环),生物素标记肽,PEG修饰肽,甲基化修饰肽等。
以上所有内容,为专肽生物原创内容,请勿发布到其他网站上。





