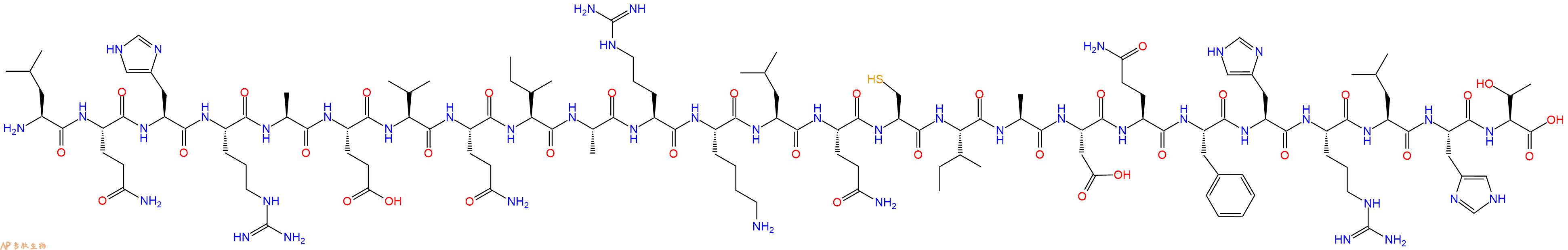
| 编号: | 408166 |
| 中文名称: | BMf-BH3 |
| 英文名: | BMf-BH3 |
| 单字母: | H2N-LQHRAEVQIARKLQCIADQFHRLHT-OH |
| 三字母: | H2N N端氨基 -Leu亮氨酸 -Gln谷氨酰胺 -His组氨酸 -Arg精氨酸 -Ala丙氨酸 -Glu谷氨酸 -Val缬氨酸 -Gln谷氨酰胺 -Ile异亮氨酸 -Ala丙氨酸 -Arg精氨酸 -Lys赖氨酸 -Leu亮氨酸 -Gln谷氨酰胺 -Cys半胱氨酸 -Ile异亮氨酸 -Ala丙氨酸 -Asp天冬氨酸 -Gln谷氨酰胺 -Phe苯丙氨酸 -His组氨酸 -Arg精氨酸 -Leu亮氨酸 -His组氨酸 -Thr苏氨酸 -OHC端羧基 |
| 氨基酸个数: | 25 |
| 分子式: | C131H215N45O35S1 |
| 平均分子量: | 3012.45 |
| 精确分子量: | 3010.61 |
| 等电点(PI): | 12.23 |
| pH=7.0时的净电荷数: | 4.65 |
| 平均亲水性: | 0.056 |
| 疏水性值: | -0.54 |
| 消光系数: | - |
| 标签: | 细胞凋亡肽(Apoptosis Peptides) |
Definition
Apoptosis or programmed cell death is a normal component of the development and health of multicellular organisms. Cells die in response to a variety of stimuli and during apoptosis they do so in a controlled, regulated fashion.
Discovery
In 1885, Flemming W described the process of programmed cell death. John Kerr's discovery, in late 1960s, initially called "shrinkage necrosis" but which he later renamed "apoptosis", came about when his attention was caught by a curious form of liver cell death during his studies of acute liver injury in rats 1,2. Kerr in 1972 proposed the term apoptosis is for mechanism of controlled cell deletion, which appears to play a complementary but opposite role to mitosis in the regulation of animal cell populations. Its morphological features suggest that it is an active, inherently programmed phenomenon, and it has been shown that it can be initiated or inhibited by a variety of environmental stimuli, both physiological and pathological 3.
Structural Characteristics
Heterodimerization between members of the Bcl-2 family of proteins is a key event in the regulation of programmed cell death. The molecular basis for heterodimer formation was investigated by determination of the solution structure of a complex between the survival protein Bcl-xL and the death-promoting region of the Bcl-2-related protein Bak. The structure and binding affinities of mutant Bak peptides indicate that the Bak peptide adopts an amphipathic helix that interacts with Bcl-xL through hydrophobic and electrostatic interactions. Mutations in full-length Bak that disrupt either type of interaction inhibit the ability of Bak to heterodimerize with Bcl-xL 4.
The structure of the 16–amino acid peptide complexed with a biologically active deletion mutant of Bcl-xL was determined by nuclear magnetic resonance spectroscopy (NMR). The structure was determined from a total of 2813 NMR-derived restraints and is well defined by the NMR data. The Bak peptide forms a helix when complexed to Bcl-xL. The COOH terminal portion of the Bak peptide interacts predominantly with residues in the BH2 and BH3 regions. Melanoma inhibitor of apoptosis (ML-IAP) is a potent anti-apoptotic protein that is upregulated in a number of melanoma cell lines but not expressed in most normal adult tissues. Overexpression of IAP proteins, such as ML-IAP or the ubiquitously expressed X-chromosome-linked IAP (XIAP), in human cancers has been shown to suppress apoptosis induced by a variety of stimuli. X-ray crystal structures of ML-IAP-BIR in complex with Smac- and phage-derived peptides, together with peptide structure−activity-relationship data, indicate that the peptides can be modified to provide increased binding affinity and selectivity for ML-IAP-BIR relative to XIAP-BIR3 5.
Mode of Action
Upon receiving specific signals instructing the cells to undergo apoptosis a number of distinctive changes occur in the cell. Families of proteins known as caspases are typically activated in the early stages of apoptosis. These proteins breakdown or cleave key cellular components that are required for normal cellular function including structural proteins in the cytoskeleton and nuclear proteins such as DNA repair enzymes. The caspases can also activate other degradative enzymes such as DNases, which begin to cleave the DNA in the nucleus.
Apoptotic cells display distinctive morphology during the apoptotic process. Typically, the cell begins to shrink following the cleavage of lamins and actin filaments in the cytoskeleton. The breakdown of chromatin in the nucleus often leads to nuclear condensation and in many cases the nuclei of apoptotic cells take on a "horse-shoe" like appearance. Cells continue to shrink, packaging themselves into a form that allows for their removal by macrophages. There are a number of mechanisms through which apoptosis can be induced in cells. The sensitivity of cells to any of these stimuli can vary depending on a number of factors such as the expression of pro- and anti-apoptotic proteins (eg. the Bcl-2 proteins or the Inhibitor of Apoptosis Proteins), the severity of the stimulus and the stage of the cell cycle. The Bcl-2 family of proteins plays a central role in the regulation of apoptotic cell death induced by a wide variety of stimuli. Some proteins within this family, including Bcl-2 and Bcl-xL, inhibit programmed cell death, and others, such as Bax and Bak, can promote apoptosis 6, 7.
Functions
For development, Apoptosis is as needed for proper development as mitosis is. Examples: The resorption of the tadpole tail at the time of its metamorphosis into a frog occurs by apoptosis.
Integrity of the organism, Apoptosis is needed to destroy cells that represent a threat to the integrity of the organism. Examples: Cells infected with viruses8.
Cells of the immune system, as cell-mediated immune responses wane, the effector cells must be removed to prevent them from attacking body constituents. CTLs induce apoptosis in each other and even in themselves 9.
Cells with DNA damage, damage to its genome can cause a cell to disrupt proper embryonic development leading to birth defects to become cancerous.
References
1. Kerr JF (1965). A histochemical study of hypertrophy and ischaemic injury of rat liver with special reference to changes in lysosomes. Journal of Pathology and Bacteriology, 90(90):419-435.
2. Kerr JF, Wyllie AH, Currie AR (1972). Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer., 26(4):239-257.
3. O'Rourke MG, Ellem KA (2000). John Kerr and apoptosis. Med. J. Aust., 173(11-12): 616-617.
4. Franklin MC, Kadkhodayan S, Ackerly H, Alexandru D, Distefano MD, Elliott LO, Flygare JA, Mausisa G, Okawa DC, Ong D, Vucic D, Deshayes K, Fairbrother WJ (2003). Structure and function analysis of peptide antagonists of melanoma inhibitor of apoptosis (ML-IAP). Biochemistry, 42(27):8223-8231.
5. Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW (1997). Structure of bcl-xl-bak peptide complex: recognition between regulators of apoptosis. Science, 275(5302):983-986.
6. Hanada M, Aimé-Sempé C, Sato T, Reed JC (1995). Structure-function analysis of Bcl-2 protein. Identification of conserved domains important for homodimerization with Bcl-2 and heterodimerization with Bax. J. Biol. Chem., 270(20):11962-11969.
7. Cheng EHY, Levine B, Boise LH, Thompson CB, Hardwic JM (1996). Bax-independent inhibition of apoptosis by Bcl-xL.Nature, 379:554-556.
8. Alimonti JB, Ball TB, Fowke KR (2003). Mechanisms of CD4+ T lymphocyte cell death in human immunodeficiency virus infection and AIDS. J Gen Virology., 84(84): 1649-1661.
9. Werlen G, Hausmann B, Naeher D, Palmer E (2003). Signaling life and death in the thymus: timing is everything. Science. 299(5614):1859-1863.
多肽H2N-Leu-Gln-His-Arg-Ala-Glu-Val-Gln-Ile-Ala-Arg-Lys-Leu-Gln-Cys-Ile-Ala-Asp-Gln-Phe-His-Arg-Leu-His-Thr-COOH的合成步骤:
1、合成CTC树脂:称取0.63g CTC Resin(如初始取代度约为0.74mmol/g)和0.56mmol Fmoc-Thr(tBu)-OH于反应器中,加入适量DCM溶解氨基酸(需要注意,此时CTC树脂体积会增大好几倍,避免DCM溶液过少),再加入1.4mmol DIPEA(Mw:129.1,d:0.740g/ml),反应2-3小时后,可不抽滤溶液,直接加入1ml的HPLC级甲醇,封端半小时。依次用DMF洗涤2次,甲醇洗涤1次,DCM洗涤一次,甲醇洗涤一次,DCM洗涤一次,DMF洗涤2次(这里使用甲醇和DCM交替洗涤,是为了更好地去除其他溶质,有利于后续反应)。得到 Fmoc-Thr(tBu)-CTC Resin。结构图如下:
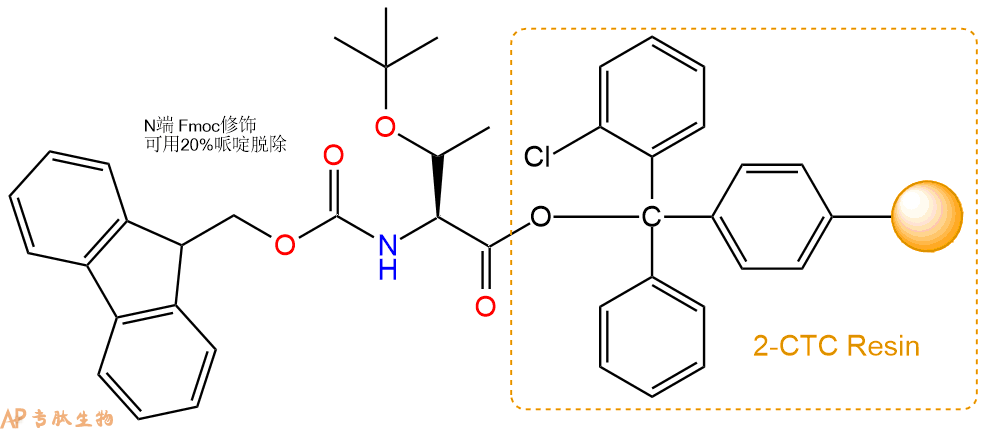
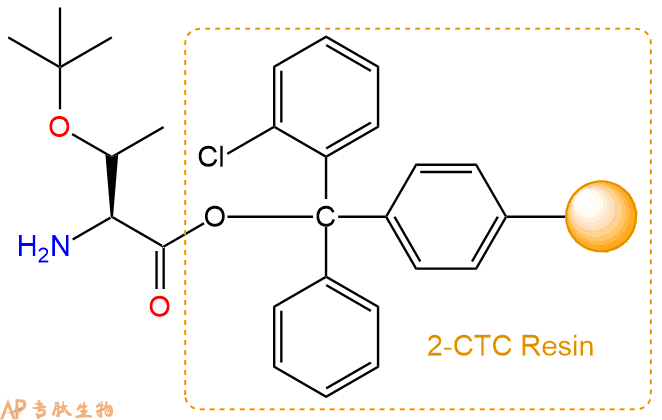
2、脱Fmoc:加3倍树脂体积的20%Pip/DMF溶液,鼓氮气30分钟,然后2倍树脂体积的DMF 洗涤5次。得到 H2N-Thr(tBu)-CTC Resin 。(此步骤脱除Fmoc基团,茚三酮检测为蓝色,Pip为哌啶)。结构图如下:
3、缩合:取1.4mmol Fmoc-His(Trt)-OH 氨基酸,加入到上述树脂里,加适当DMF溶解氨基酸,再依次加入2.8mmol DIPEA,1.33mmol HBTU。反应30分钟后,取小样洗涤,茚三酮检测为无色。用2倍树脂体积的DMF 洗涤3次树脂。(洗涤树脂,去掉残留溶剂,为下一步反应做准备)。得到Fmoc-His(Trt)-Thr(tBu)-CTC Resin。氨基酸:DIPEA:HBTU:树脂=3:6:2.85:1(摩尔比)。结构图如下:

4、依次循环步骤二、步骤三,依次得到
H2N-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Ala-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Ala-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Arg(Pbf)-Ala-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Arg(Pbf)-Ala-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-His(Trt)-Arg(Pbf)-Ala-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-His(Trt)-Arg(Pbf)-Ala-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Gln(Trt)-His(Trt)-Arg(Pbf)-Ala-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
H2N-Gln(Trt)-His(Trt)-Arg(Pbf)-Ala-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
Fmoc-Leu-Gln(Trt)-His(Trt)-Arg(Pbf)-Ala-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin
以上中间结构,均可在专肽生物多肽计算器-多肽结构计算器中,一键画出。
最后再经过步骤二得到 H2N-Leu-Gln(Trt)-His(Trt)-Arg(Pbf)-Ala-Glu(OtBu)-Val-Gln(Trt)-Ile-Ala-Arg(Pbf)-Lys(Boc)-Leu-Gln(Trt)-Cys(Trt)-Ile-Ala-Asp(OtBu)-Gln(Trt)-Phe-His(Trt)-Arg(Pbf)-Leu-His(Trt)-Thr(tBu)-CTC Resin,结构如下:
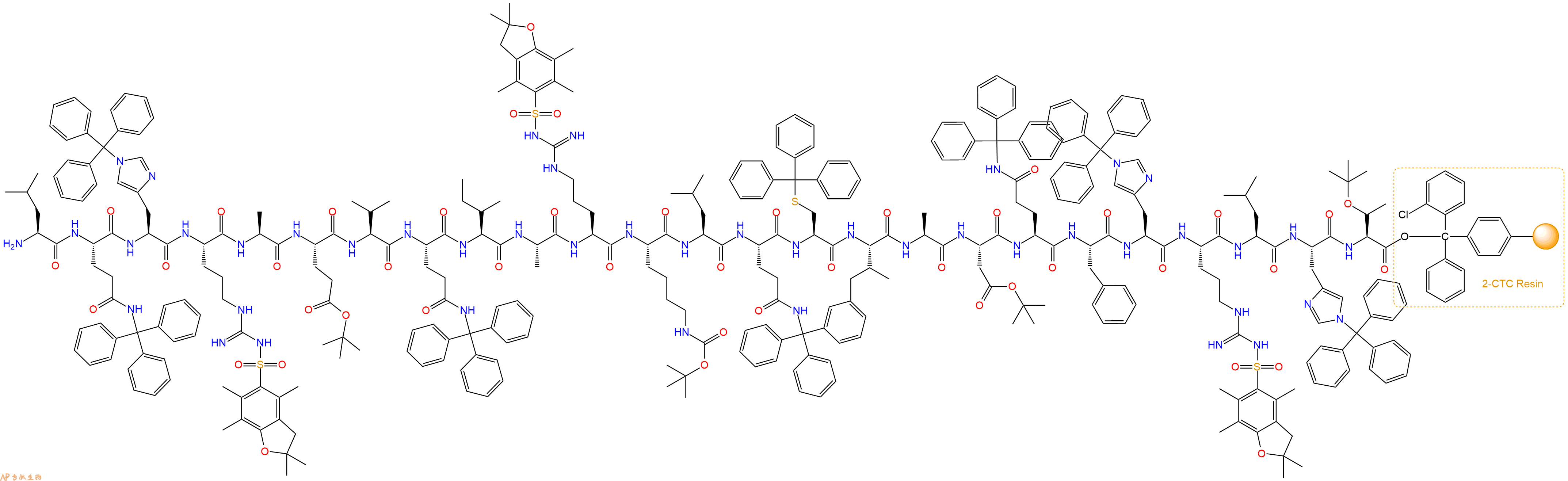
5、切割:6倍树脂体积的切割液(或每1g树脂加8ml左右的切割液),摇床摇晃 2小时,过滤掉树脂,用冰无水乙醚沉淀滤液,并用冰无水乙醚洗涤沉淀物3次,最后将沉淀物放真空干燥釜中,常温干燥24小试,得到粗品H2N-Leu-Gln-His-Arg-Ala-Glu-Val-Gln-Ile-Ala-Arg-Lys-Leu-Gln-Cys-Ile-Ala-Asp-Gln-Phe-His-Arg-Leu-His-Thr-COOH。结构图见产品结构图。
切割液选择:1)TFA:H2O=95%:5%
2)TFA:H2O:TIS=95%:2.5%:2.5%
3)三氟乙酸:茴香硫醚:1,2-乙二硫醇:苯酚:水=87.5%:5%:2.5%:2.5%:2.5%
(前两种适合没有容易氧化的氨基酸,例如Trp、Cys、Met。第三种适合几乎所有的序列。)
6、纯化冻干:使用液相色谱纯化,收集目标峰液体,进行冻干,获得蓬松的粉末状固体多肽。不过这时要取小样复测下纯度 是否目标纯度。
7、最后总结:
杭州专肽生物技术有限公司(ALLPEPTIDE https://www.allpeptide.com)主营定制多肽合成业务,提供各类长肽,短肽,环肽,提供各类修饰肽,如:荧光标记修饰(CY3、CY5、CY5.5、CY7、FAM、FITC、Rhodamine B、TAMRA等),功能基团修饰肽(叠氮、炔基、DBCO、DOTA、NOTA等),同位素标记肽(N15、C13),订书肽(Stapled Peptide),脂肪酸修饰肽(Pal、Myr、Ste),磷酸化修饰肽(P-Ser、P-Thr、P-Tyr),环肽(酰胺键环肽、一对或者多对二硫键环),生物素标记肽,PEG修饰肽,甲基化修饰肽等。
以上所有内容,为专肽生物原创内容,请勿发布到其他网站上。





